There many types of RNA in a cell Nevertheless, we will focus on the three that are straight related to producing proteins: messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). DNA can not produce proteins without their help. The other RNA types play various regulative roles that we will dismiss.
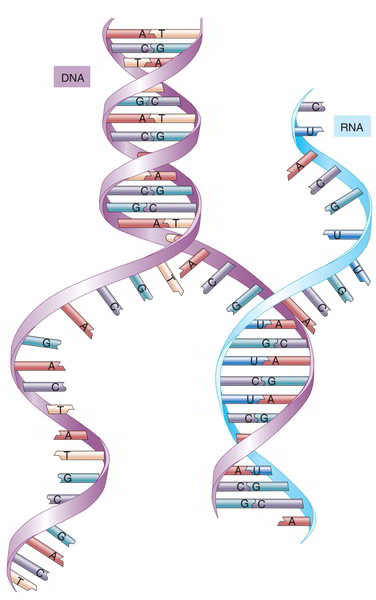
The most significant difference is that RNA is much smaller sized, differing from about 70 to 90 bases in tRNA to rather over 10,000bases in the most important mRNA. DNA, by contrast, averages more than 100 million base sets long Similarly, whereas DNA is a double helix, RNA consists of simply one nucleotide chain, not held together by complementary base sets besides in specific quick areas where the molecule folds back on itself. The sugar in RNA is ribose rather of deoxyribose. RNA includes 3 of the precise very same nitrogenous bases as DNA– adenine, cytosine, and guanine– nevertheless it has no thymine; a base called uracil (U) takes its place Transfer RNA has more than 50 numerous nitrogenous bases, however, for our functions, we do not need to think of other than A, U, C, and G.
The essential role of the three main RNAs is to translate the code in DNA and make use of those instructions to synthesise proteins RNA is a non-reusable molecule that usually works in the cytoplasm, while DNA is irreplaceable and remains firmly behind in the nucleus, “offering orders” from there.
RNA Synthesis
Each gene is a stretch of DNA that is a number of thousand nucleotide sets long The DNA in a human cell includes over 3 billion base sets– sufficient to code for a minimum of 3 million proteins Due to the fact that the typical human cell contains fewer proteins than this (30,000to 150,000various proteins), it follows that just a portion of the DNA in each cell is utilized to code for proteins A few of the DNA might be non-active or redundant, and some serves to control those regions that do code for proteins.
For the genetic code to be equated into the synthesis of particular proteins, the DNA code initially should be copied onto a strand of RNA. This is achieved by DNA-directed RNA synthesis– the process of genetic transcription.
There are base series for “start” and “stop,” and regions of DNA that work as promoters of gene transcription. Numerous regulative molecules, such as some hormones, function as transcription aspects by binding to the promoter region of a particular gene and stimulating genetic transcription. Transcription (RNA synthesis) needs the enzyme RNA polymerase, which engages with a promoter region to transcribe a persongene This enzyme has a globular structure with a big central cavity; when it breaks the hydrogen bonds between DNA strands, the apart strands are required apart within this cavity The released bases can then match (by hydrogen bonding) with complementary RNA nucleotide bases present in the nucleoplasm.
This pairing of bases, like that which takes place in DNA replication (explained in a later area), follows the law of complementary base pairing: guanine bonds with cytosine (and vice versa), and adenine bonds with uracil (since uracil in RNA is comparable to thymine in DNA). Unlike DNA replication, nevertheless, just one of the two released strands of DNA functions as a guide for RNA synthesis. When an RNA molecule has been produced, it removes from the DNA strand on which it was formed. This process can continue forever, providing numerous countless RNA copies of the DNA strand that is being transcribed. When the gene is not to be transcribed, the apart DNA strands can then return together once again.
Types of RNA
There are 4 types of RNA needed for gene expression: (1) precursor messenger RNA (pre-mRNA), which is changed within the nucleus to form mRNA; (2) messenger RNA (mRNA), which includes the code for the synthesis of
particular proteins; (3) transfer RNA (tRNA), which is required for deciphering the genetic message contained in mRNA; and (4) ribosomal RNA (rRNA), which forms part of the structure of ribosomes. The DNA that codes for rRNA synthesis lies in the part of the nucleus called the nucleolus. The DNA that codes for pre-mRNA and tRNA synthesis lies in other places in the nucleus.
In bacteria, where the molecular biology of the gene is finest comprehended, a gene that codes for one type of protein produce an mRNA molecule that starts to direct protein synthesis as quickly as it is transcribed. This is not the case in greater organisms, consisting of human beings. In greater cells, a pre-mRNA is produced that should be customised within the nucleus before it can go into the cytoplasm as mRNA and direct protein synthesis.
Precursor mRNA is much bigger than the mRNA it forms. Remarkably, this plus size of pre-mRNA is not due to excess bases at the ends of the molecule that should be cut; rather, the excess bases lie within the pre-mRNA. The genetic code for a particular protein, to puts it simply, is broken up by stretches of base sets that do not add to the code. These regions of noncoding DNA within a gene are called introns; the coding regions are called exons. As a result, pre-mRNA should be cut and entwined to make mRNA.
When the human genome was sequenced, and it was found that we have about 25,000genes but produce more than 100,000various proteins, it ended up being clear that a person gene might code for more than one protein Certainly, specific genes code for approximately 3 various proteins To a big degree, this is achieved by alternative splicing of exons. Depending upon which lengths of the gene’s base sets are eliminated as introns and which work as exons to be entwined together, an offered gene can produce a number of various mRNA molecules, coding for a number of various proteins.
An approximated 92 % to 94 % of human genes go through alternative splicing of exons, with the majority of the variation happening betweenvarioustissues The average gene includes eight exons, although the number can be much bigger– the gene for the protein “titin” includes 234 exons! Entwining together these exons in various methods might produce numerous variations of the protein product The human proteome is therefore much bigger than the genome, enabling remarkable versatility for various functions.
Introns are eliminated of the pre-mRNA, and completions of the exons are entwined, by macromolecules called snRNPs (noticeable “snurps”), producing the functional mRNA that leaves the nucleus and goes into the cytoplasm. SnRNPs mean little nuclear ribonucleoproteins. These are little, ribosome-like aggregates of RNA and protein that form a body called a spliceo-some that entwines the exons together.
Do the introns– eliminated from pre-mRNA in the formation of mRNA– have a functional significance? And, because less than 2 % of the DNA codes for proteins, exactly what about all the other DNA situated between the protein- coding genes? Is everything “scrap”? Researchers as soon as believed so, however proof recommends that RNA molecules can themselves have essential regulative functions in the cell. For instance, in many cases, the RNA transcribed from regions of DNA that do not code for proteins has actually been revealed to assist control the expression of regions that do. This suggests that a description of the genome, as well as of the proteome, might not supply a total understanding of all the manner ins which DNA controls the cell.
RNA Interference
The 2006 Nobel Reward in Physiology or Medication was granted for the discovery of RNA disturbance (RNA.), a regulative process carried out by RNAmolecules In this process, specific RNA molecules that do not code for proteins might avoid particular mRNA molecules from being revealed (equated). RNA disturbance is moderated by two extremely comparable types of RNA. One type is formed from longer double-stranded RNA molecules that leave the nucleus and are processed in the cytoplasm by an enzyme (called Dicer) into brief (21to 25 nucleotides long) double-stranded RNA molecules called brief interfering RNA, or siRNA. The double-stranded RNA is formed from either the transcription of a sector of 2 complementary DNA strands, or from double-stranded RNA produced by a virus inside the host cell. In this, RNA interference is a mechanism to assist fight the viral infection.
The other type of brief RNA that takes part in RNA interference is formed from longer RNA strands that fold into barrette loops that look like double-stranded RNA. These are processed by an enzyme in the nucleus then Dicer in the cytoplasm into brief (about 23 nucleotides long) double-stranded RNA molecules called microRNA (miRNA). Among the 2 strands from the siRNA and miRNA then go into a protein particle called the RNA-induced silencing complex (RISC), so that this single- stranded RNA can match by complementary base bonding to particular mRNA molecules targeted for interference.
There can be a range in the degree of complementary base pairings between one siRNA or miRNA and a variety of various mRNAs. An siRNA can be completely complementary to a specific mRNA, forming an siRNA-mRNA duplex. In this case, the RISC will avoid the mRNA from being equated by triggering the destruction of the mRNA. As a result, a single siRNA can silence one specific mRNA. A lot of miRNA are not adequately complementary to the mRNA to cause the mRNA’s destruction; rather, the miRNA avoids the mRNA from being equated intoprotein We have numerous unique miRNA genes that control the expression of an even higher number of mRNAgenes This is possible since one miRNA can be incompletely complementary to a variety of various mRNA molecules (from various genes), triggering them to be silenced. In this method, a single miRNA might silence as numerous as an approximated 200 various mRNAmolecules Researchers presently approximate that a minimum of 30% of human genes are controlled by miRNAs.
Researchers have actually found a couple of hundred various miRNA molecules in human beings and have actually created libraries of miRNAs to silence the expression of numerous genes This can assist in the study of typical genetic regulation and might cause medical applications. For instance, an miRNA that prevents expression of a growth suppressor gene can promote cancer, whereas a various miRNA that quelches an oncogene (which promotes cancer) might have the opposite result. In basic, growth cells produce less miRNA molecules than typical cells, and modifications in the miRNA profile of metastatic cancer may be utilised to figure out the origin, aggressiveness, and the majority of efficient treatment
of the cancer. A specific miRNA that reduces the expression of cyclin proteins, required for development through the cell cycle (gone over in area 3.5), was just recently discovered to be unusually decreased in mouse liver cancer cells; the intro of this miRNA into the growth cells prevented their expansion and the growth of this cancer.
In the future, RNA disturbance might be utilised clinically to reduce the expression of particular genes, either unusual genes of the client or the genes of transmittable infections. At the time of this writing, making use of an siRNA to deal with age-related macular degeneration (a significant reason for loss of sight) remains in phase III clinical trials, and others remain in development to treat this very same disease along with the respiratory syncytial virus, high blood cholesterol, Huntington’s disease, hepatitis C, strong growths, AIDS lymphoma, and other conditions. Additionally, drugs in development to deal with hepatitis C and other conditions are developed to obstruct the capability of particular miRNA molecules to prevent genetic expression. Although these drugs might show efficient, their security is a continuing issue.

 (51 votes, average: 4.59 out of 5)
(51 votes, average: 4.59 out of 5)